Loop Assembly
Loop Assembly has been developed in the Haseloff Lab by Bernardo Pollak and Fernarn Federici, is an efficient and simple type IIS method that uses BsaI and SapI as type IIS enzymes, and two sets of 4 plasmids for cloning from one level into the next. This is the method being used in Marchantia to assemble L1 and L2 constructs. Paper on Loop available in bioRxiv. More details here.
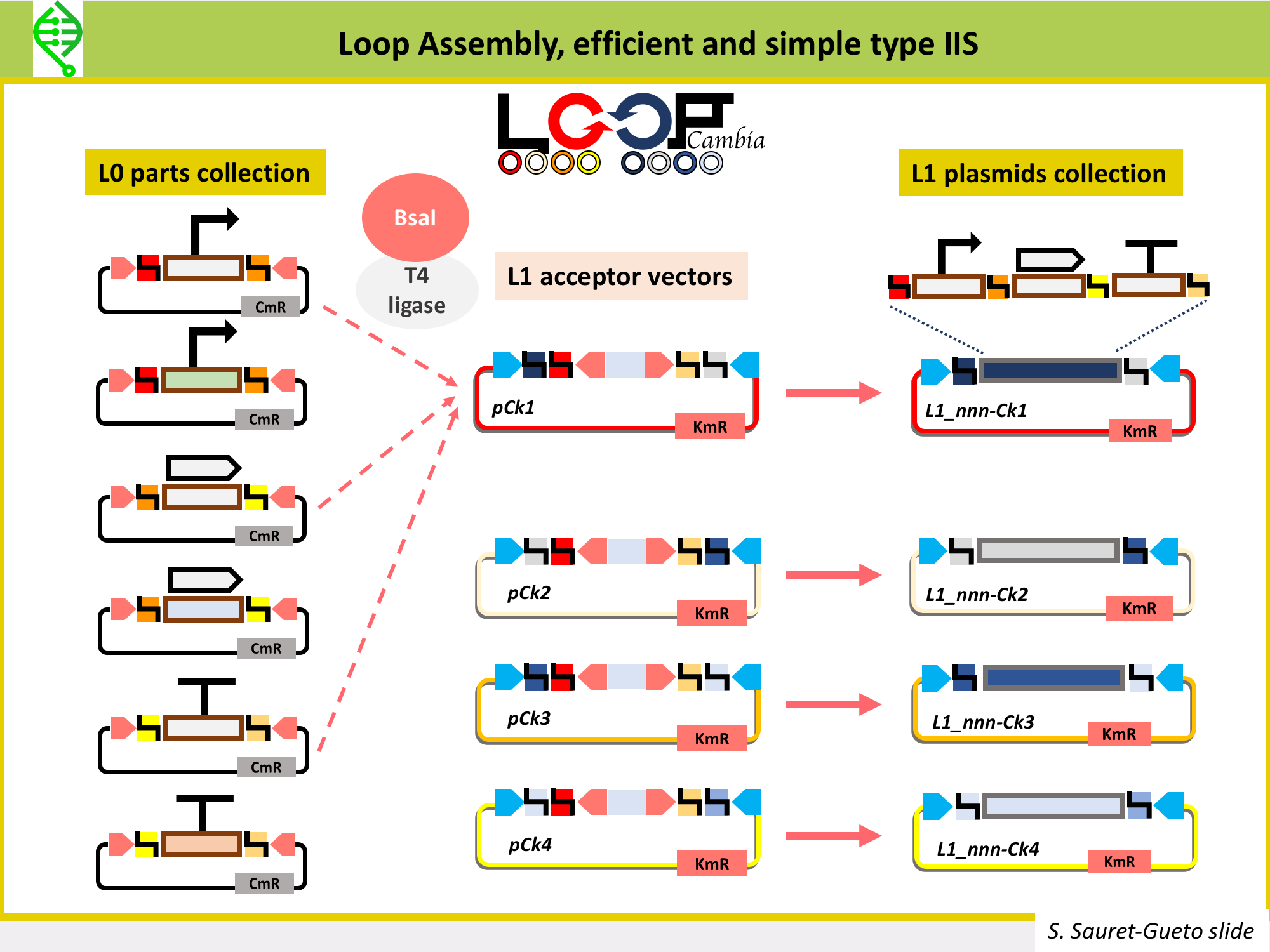
Also, in the Haaseloff Lab Eftychios Frangedakis, Marta Tomaselli and Susana Sauret-Gueto in collaboration with Anthony West and Nicola Patron at the Erlham Institute in Norwich, have developed Loop-Cambia, with same framework as Loop but different set of cloning vectors, which are higher copy number and more efficient for all types of Loop.
Loop Assembly
Developed in Jim Haseloff Lab by Bernardo Pollack and Fernan Federici, the manuscript is already available in bioRxiv.
Odd receivers:
pOd1, pOd2, pOd3, pOd4. Low copy number pGreen-based plasmids. Confer KanR . Cloning into with BsaI. To assemble L1 contructs (and L3, L5...). Cloning out with SapI.
EVEN receivers:
pEv1, pEv2, pEv3, pEv4. Low copy number pGreen-based plasmids. Confer SepcR. Cloning into with SapI. To assemble L2 contructs (and L4, L6..). Cloning out with BsaI.
Loop-Cambia
Continuation of Loop by Susana Sauret-Gueto, Eftychios Frangedakis, Anthonty West, Nicola Patron, Jim Haseloff.
L1 receivers:
pCk1, pCk2, pCk3, pCk4. Higher copy number pCambia-based plasmids. Confer KanR. Cloning into with BsaI. To assemble L1 contructs (and L3, L5...). Cloning out with SapI
L2 receivers:
pCsA, pCsB, pCsC, pCsE. Higher copy number pCambia-based plasmids. Confer SpecR. Cloning into with SapI. To assemble L2 contructs (and L4, L6..). Cloning out with BsaI.